![]()
In this case the Z-axis lies along a line joining the centres of the hexagonal faces.
The Y-axis is perpendicular to this and passes through the centre of one of the faces. The
X-axis completes the orthonormal set and passes through the centre of an edge that is
parallel to the Z-axis. (Note: It is important to get this convention right!) The origin
of the atomic coordinates is the centre of the cell. If the length of one of the hexagon
edges is D, the cell vectors required in the CONFIG file are: (3D,0,0), (0,![]() D,0), (0,0,H),
where H is the prism height (the distance between hexagonal faces). The orthorhombic cell
also defined by these vectors enscribes the hexagonal prism and possesses twice the
volume, but the height and the centre are the same.
D,0), (0,0,H),
where H is the prism height (the distance between hexagonal faces). The orthorhombic cell
also defined by these vectors enscribes the hexagonal prism and possesses twice the
volume, but the height and the centre are the same.
The Ewald summation method may be used with this periodic boundary condition.
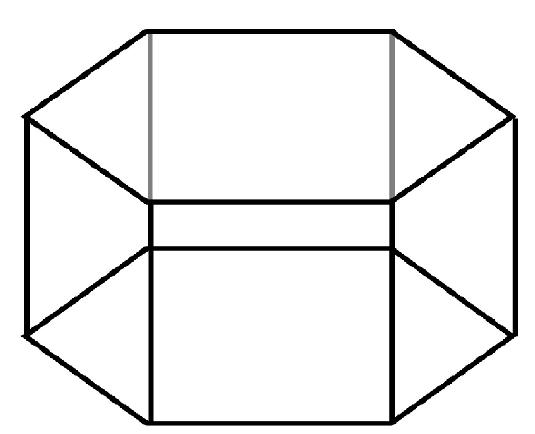
The hexagonal MD cell.
This MD cell is particularly suitable for simulating strands or fibres (i.e. systems with a pronounced anisotropy in the Z-direction), such as DNA strands in solution, or stretched polymer chains.
![]()