


Next: About this document ...
Up: THE DL_POLY_2 USER MANUAL
Previous: Message 514: error -
Contents
- algorithm
- no title
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Integration algorithms
| The CONTROL File
- Brode-Ahlrichs
- The DL_POLY_2 Force Field
| The Replicated Data Strategy
| Distributing the Nonbonded Terms
| Distributing the Nonbonded Terms
- FIQA
- Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration of the Rigid
- multiple timestep
- The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| Distributing the Nonbonded Terms
| Constructing Nonstandard Versions
| Constructing Nonstandard Versions
| The CONTROL File Directives
| The CONTROL File Directives
| Further Comments on the
| Further Comments on the
| Further Comments on the
| Simulation Progress
| Test Case 1: KNaSi
 O
O | Message 153: error -
| Message 385: error -
| Message 385: error -
| Message 386: error -
| Message 398: error -
| Message 422: error -
| Message 153: error -
| Message 385: error -
| Message 385: error -
| Message 386: error -
| Message 398: error -
| Message 422: error -
- QSHAKE
- Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Linked Rigid Bodies
| Linked Rigid Bodies
| Linked Rigid Bodies
| Linked Rigid Bodies
| The RD-SHAKE and Parallel
- RD-SHAKE
- Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Bond Constraints
| The RD-SHAKE and Parallel
| The RD-SHAKE and Parallel
| Message 102: error -
| Message 105: error -
| Message 190: error -
- SHAKE
- Molecular Dynamics Algorithms
| The Replicated Data Strategy
| The RD-SHAKE and Parallel
| The RD-SHAKE and Parallel
| The RD-SHAKE and Parallel
| The RD-SHAKE and Parallel
| The Parameters and their
| Message 438: error -
- Verlet
- Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Short Ranged (van der
| Atomistic and Charge Group
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Bond Constraints
| Bond Constraints
| Bond Constraints
| Bond Constraints
| Bond Constraints
| Nosé- Hoover Thermostat
| Gaussian Constraints
| The Hoover Barostat
| The RD-SHAKE and Parallel
- AMBER
- The DL_POLY Package
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Macromolecules
| Macromolecules
| Adding Solvent to a
- angular restraints
- Angular Restraints
- barostat
- Molecular Dynamics Algorithms
| Integration of the Rigid
| Integration of the Rigid
| The CONTROL File Directives
| Message 466: error -
- Berendsen
- Berendsen Barostat
| Linked Rigid Bodies
| Test Case 5: Shell
- Hoover
- The Hoover Barostat
| The Hoover Barostat
- boundary conditions
- Boundary Conditions
| Boundary Conditions
| Long Ranged Electrostatic (Coulombic)
| Constructing Nonstandard Versions
| Inorganic Materials
| Test Case 3: An
| Test Case 5: Shell
| Test Case 10: DNA
| Introduction
| Introduction
| Message 67: error -
| no title
- cubic
- Further Comments
| Message 67: error -
| Message 79: error -
| Message 300: error -
- hexagonal prism
- Further Comments
- parallelpiped
- Message 67: error -
| Message 79: error -
| Message 300: error -
- rhombic dodecahedron
- Further Comments
- slab
- Message 300: error -
- truncated octahedron
- Further Comments
- CCP5
- The DL_POLY Package
| The DL_POLY Package
| The DL_POLY Package
| The DL_POLY Package
| The public Sub-directory
| Obtaining the Source Code
- charge groups
- .
| Test Case 4: Shell
| Message 97: error -
| Message 220: error -
| Message 230: error -
- COMMON blocks
- Internal Data Transfer
| Internal Data Transfer
| FORTRAN Parameters
| Modifying the Makefile
- constrains
- bond
- The Parameters and their
- constraints
- bond
- Molecular Systems
| Molecular Systems
| Molecular Dynamics Algorithms
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Bond Constraints
| Bond Constraints
| Potential of Mean Force
| Potential of Mean Force
| Description of Rigid Body
| Description of Rigid Body
| Description of Rigid Body
| Integration of the Rigid
| Linked Rigid Bodies
| Linked Rigid Bodies
| Linked Rigid Bodies
| The RD-SHAKE and Parallel
| The RD-SHAKE and Parallel
| The RD-SHAKE and Parallel
| The RD-SHAKE and Parallel
| Constructing Nonstandard Versions
| Constructing Nonstandard Versions
| .
| .
| Simulation Progress
| Test Case 3: An
| Test Case 7: Linked
| Test Case 9: A
| The Parameters and their
| Message 40: error -
| Message 41: error -
| Message 70: error -
| Message 99: error -
| Message 105: error -
| Message 432: error -
- Gaussian
- Ewald Sum
| Ewald Sum
| Ewald Sum
| Ewald Sum
| Ewald Sum
| Ewald Sum
| Thermostats
| Test Case 2: Metal
- PMF
- Potential of Mean Force
| Potential of Mean Force
| .
| .
- CVS
- Version Control System (CVS)
| Version Control System (CVS)
| Internal Documentation
- direct Coulomb sum
- Long Ranged Electrostatic (Coulombic)
| Direct Coulomb Sum
| Coulomb Sum with Distance
- distance dependant dielectric
- Coulomb Sum with Distance
| Reaction Field
| The CONTROL File Directives
| Further Comments on the
| Message 424: error -
- distance restraints
- Distance Restraints
| Distance Restraints
- DLPROTEIN
- Macromolecules
- ensemble
- Molecular Dynamics Algorithms
| no title
| Message 430: error -
- Berendsen N
 T
T
- The CONTROL File Directives
- Berendsen N
 T
T
- Further Comments on the
- Berendsen N
 T
T
- Further Comments on the
- Berendsen N
 T
T
- Molecular Dynamics Algorithms
- Berendsen N
 T
T
- Molecular Dynamics Algorithms
- Berendsen N
 T
T
- Molecular Dynamics Algorithms
- Berendsen N
 T
T
- Integration algorithms
- Berendsen N
 T
T
- Integration algorithms
- Berendsen N
 T
T
- Integration algorithms
- Berendsen NPT
- Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Further Comments on the
| Further Comments on the
- Berendsen NVT
- Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| The CONTROL File Directives
| The CONTROL File Directives
| Further Comments on the
| Further Comments on the
- BerendsenN
 T
T
- Integration algorithms
- canonical
- Thermostats
- Evans NVT
- Molecular Dynamics Algorithms
| Integration algorithms
| Integration algorithms
| The CONTROL File Directives
| Further Comments on the
| Further Comments on the
- Hoover N
 T
T
- Further Comments on the
- Hoover N
 T
T
- Molecular Dynamics Algorithms
- Hoover N
 T
T
- Molecular Dynamics Algorithms
- Hoover N
 T
T
- Molecular Dynamics Algorithms
- Hoover N
 T
T
- Integration algorithms
- Hoover N
 T
T
- Integration algorithms
- Hoover N
 T
T
- Integration algorithms
- Hoover N
 T
T
- Integration algorithms
- Hoover NPT
- Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| The CONTROL File Directives
| Further Comments on the
- Hoover NVT
- Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Integration algorithms
| Further Comments on the
- microcanonical
- see ensemble,NVE
- NVE
- Thermostats
| The CONTROL File Directives
| Further Comments on the
| Further Comments on the
| Further Comments on the
- error messages
- The DL_POLY_2 Internal Error
| Introduction to Message 514: error -
- Ewald
- Hautman Klein
- Long Ranged Electrostatic (Coulombic)
| no title
| Hautman Klein Ewald Optimisation
| The CONTROL File Directives
| Slab boundary conditions (IMCON=6)
- optimisation
- Ewald sum and SPME
| Ewald sum and SPME
| Ewald sum and SPME
- SPME
- Required Program Libraries
| Long Ranged Electrostatic (Coulombic)
| Smoothed Particle Mesh Ewald
| Modifying the Makefile
| Ewald sum and SPME
| The CONTROL File Directives
| Test Case 10: DNA
- summation
- Long Ranged Electrostatic (Coulombic)
| Long Ranged Electrostatic (Coulombic)
| Atomistic and Charge Group
| Ewald Sum
| Ewald Sum
| Ewald Sum
| Ewald Sum
| Ewald Sum
| Ewald Sum
| The DL_POLY_2 Multiple Timestep
| The Replicated Data Strategy
| Modifications for the Ewald
| Ewald sum and SPME
| Ewald sum and SPME
| Ewald sum and SPME
| Ewald sum and SPME
| Ewald sum and SPME
| The CONTROL File Directives
| Further Comments on the
| Further Comments on the
| Test Case 1: KNaSi
 O
O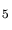 | Test Case 5: Shell
| Test Case 9: A
| Message 180: error -
| Message 185: error -
| Message 250: error -
| Message 330: error -
| Message 433: error -
| Test Case 5: Shell
| Test Case 9: A
| Message 180: error -
| Message 185: error -
| Message 250: error -
| Message 330: error -
| Message 433: error -
- force field
- no title
| The DL_POLY_2 Force Field
| no title
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Dihedral Angle Potentials
| External Fields
| External Fields
| Adding Solvent to a
| Message 12: error -
| Message 105: error -
- AMBER
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
- DL_POLY
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field to Dynamical Shell Model
- Dreiding
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Three Body Potentials
| Three Body Potentials
| Test Case 9: A
| Test Case 10: DNA
| Test Case 11: Hautman-Klein
- GROMOS
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
- FORTRAN 77
- Programming Language
| Target Computers
| Internal Documentation
| Internal Documentation
| FORTRAN Parameters
| Constructing the Standard Version
| Modifying the Makefile
| Function
- FORTRAN 90
- Programming Language
| Memory Management
| Internal Data Transfer
- FTP
- The public Sub-directory
| Obtaining the Source Code
| Obtaining the Source Code
| Obtaining the Source Code
- Graphical User Interface
- The java Sub-directory
| A Guide to Preparing
| Analysing Results
| Definitions of Variables
- GROMOS
- The DL_POLY Package
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
- Hautman Klein Ewald
- see Ewald� Hautman Klein
- Java GUI
- Java Graphical User Interface
| The java Sub-directory
- licence
- The DL_POLY Package
- long range corrections
- Sutton-Chen
- Metal Potentials
- van der Waals
- Short Ranged (van der
- parallelisation
- no title
| The Replicated Data Strategy
- Ewald summation
- Modifications for the Ewald
- intramolecular terms
- Distributing the Intramolecular Bonded
| Distributing the Intramolecular Bonded
- Replicated Data
- Parallel Algorithms
- Verlet neighbour list
- Distributing the Nonbonded Terms
- polarisation
- Dynamical Shell Model
| Dynamical Shell Model
| Test Case 5: Shell
- dynamical shell model
- Dynamical Shell Model
| Dynamical Shell Model
| Dynamical Shell Model
| Message 93: error -
| Message 97: error -
- potential
- bond
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Bond Potentials
| Bond Potentials
| Bond Potentials
| Bond Potentials
| Distance Restraints
| Dihedral Angle Potentials
| Improper Dihedral Angle Potentials
| Tethering Forces
| Three Body Potentials
| Dynamical Shell Model
| Distributing the Intramolecular Bonded
| The RD-SHAKE and Parallel
| Inorganic Materials
| .
| Simulation Progress
| The Parameters and their
| The Parameters and their
| The Parameters and their
| The Parameters and their
| Message 30: error -
| Message 31: error -
| Message 444: error -
- Coulombic
- see potential,electrostatic
- dihedral
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Dihedral Angle Potentials
| Dihedral Angle Potentials
| Dihedral Angle Potentials
| Improper Dihedral Angle Potentials
| Improper Dihedral Angle Potentials
| Improper Dihedral Angle Potentials
| Improper Dihedral Angle Potentials
| Improper Dihedral Angle Potentials
| Inversion Angle Potentials
| The Replicated Data Strategy
| Distributing the Intramolecular Bonded
| .
| Simulation Progress
| The Parameters and their
| The Parameters and their
| The Parameters and their
| Message 60: error -
| Message 61: error -
| Message 448: error -
- electrostatic
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Units
| The DL_POLY_2 Force Field
| Distance Restraints
| Angular Restraints
| no title
| Long Ranged Electrostatic (Coulombic)
| Atomistic and Charge Group
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The CONTROL File Directives
| The CONTROL File Directives
| The CONTROL File Directives
| The CONTROL File Directives
| Further Comments on the
| Simulation Progress
| Simulation Progress
| Test Case 1: KNaSi
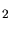 O
O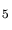 | Test Case 3: An
| Test Case 9: A
| Message 383: error -
| Message 424: error -
| Message 446: error -
| Test Case 3: An
| Test Case 9: A
| Message 383: error -
| Message 424: error -
| Message 446: error -
- four-body
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The Intermolecular Potential Functions
| Four Body Potentials
| Four Body Potentials
| Four Body Potentials
| Four Body Potentials
| Four Body Potentials
| Four Body Potentials
| Four Body Potentials
| .
| .
| .
| Simulation Progress
| The Parameters and their
| The Parameters and their
| Message 19: error -
| Message 79: error -
| Message 89: error -
| Message 91: error -
| no title
| Message 443: error -
| Message 453: error -
- improper dihedral
- The DL_POLY_2 Force Field
| Improper Dihedral Angle Potentials
| Improper Dihedral Angle Potentials
| The Replicated Data Strategy
- intramolecular
- The Intermolecular Potential Functions
| Four Body Potentials
| Constructing Nonstandard Versions
- inversion
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Inversion Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Four Body Potentials
| Four Body Potentials
| .
| The Parameters and their
| The Parameters and their
| The Parameters and their
| The Parameters and their
| no title
| Message 449: error -
- metal
- The DL_POLY_2 Force Field
| Metal Potentials
| Message 71: error -
| Message 71: error -
| Message 151: error -
- nonbonded
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Distributing the Nonbonded Terms
| Distributing the Nonbonded Terms
| Three Body Forces
| Note on Interpolation Schemes
| Note on Interpolation Schemes
| Inorganic Materials
| Adding Solvent to a
| The CONTROL File Directives
| Definitions of Variables
| .
| .
| Non-bonded Interactions
| Message 13: error -
| Message 13: error -
- Sutton-Chen
- see potential,metal
- tabulated
- The TABLE File
| Message 22: error -
| Message 23: error -
- tethered
- Tethering Forces
| Tethering Forces
| Tethering Forces
| Tethering Forces
| Frozen Atoms
| .
| Simulation Progress
| Simulation Progress
| Test Case 8: An
| The Parameters and their
| The Parameters and their
| no title
| Message 450: error -
- three-body
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Valence Angle Potentials
| The Intermolecular Potential Functions
| Three Body Potentials
| Three Body Potentials
| Three Body Potentials
| Three Body Potentials
| Three Body Potentials
| Three Body Potentials
| Three Body Potentials
| Three Body Forces
| Inorganic Materials
| Inorganic Materials
| .
| .
| .
| Simulation Progress
| Simulation Progress
| The Parameters and their
| The Parameters and their
| Message 18: error -
| Message 69: error -
| no title
| Message 84: error -
| no title
| Message 86: error -
| Message 451: error -
- torsion
- see potential,dihedral
- valence angle
- The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| The DL_POLY_2 Force Field
| Valence Angle Potentials
| Valence Angle Potentials
| Valence Angle Potentials
| Inversion Angle Potentials
| Three Body Potentials
| Three Body Potentials
| Three Body Potentials
| The Replicated Data Strategy
| Three Body Forces
| Constructing Nonstandard Versions
| Inorganic Materials
| .
| .
| Simulation Progress
| The Parameters and their
| The Parameters and their
| The Parameters and their
| The Parameters and their
| Message 50: error -
| Message 51: error -
- van der Waals
- The DL_POLY_2 Force Field
| Distance Restraints
| Angular Restraints
| The DL_POLY_2 Multiple Timestep
| Constructing Nonstandard Versions
| Note on Interpolation Schemes
| Further Comments on the
| .
| The Parameters and their
| Message 400: error -
| Message 402: error -
- quaternions
- Molecular Dynamics Algorithms
| Integration algorithms
| Integration of the Rigid
| Integration of the Rigid
| The CONTROL File Directives
- reaction field
- no title
| Reaction Field
| Reaction Field
- reactionfield
- The CONTROL File Directives
- respa
- Molecular Dynamics Algorithms
| The respa Sub-directory
| The DL_POLY_2 RESPA Multiple
- rigid body
- Molecular Systems
| Molecular Systems
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Molecular Dynamics Algorithms
| Frozen Atoms
| Integration algorithms
| Bond Constraints
| Description of Rigid Body
| Description of Rigid Body
| Integration of the Rigid
| Integration of the Rigid
| Linked Rigid Bodies
| Linked Rigid Bodies
| Linked Rigid Bodies
| Linked Rigid Bodies
| Linked Rigid Bodies
| Linked Rigid Bodies
| Linked Rigid Bodies
| Linked Rigid Bodies
| The DL_POLY_2 RESPA Multiple
| The RD-SHAKE and Parallel
| Constructing Nonstandard Versions
| Test Case 3: An
| Test Case 4: Shell
| Test Case 7: Linked
| Test Case 9: A
| The Parameters and their
| The Parameters and their
| The Parameters and their
| Message 93: error -
| Message 301: error -
| Message 302: error -
| Message 303: error -
| Message 304: error -
| Message 320: error -
| no title
| Message 360: error -
| Message 430: error -
| Message 456: error -
| Message 508: error -
- rigid bond
- see constraints,bond
- rigid ion
- Molecular Dynamics Algorithms
| The DL_POLY_2 RESPA Multiple
- rigid molecule
- see rigid body
- SPME
- see Ewald�SPME
- stress tensor
- Valence Angle Potentials
| Valence Angle Potentials
| Dihedral Angle Potentials
| Dihedral Angle Potentials
| Inversion Angle Potentials
| Inversion Angle Potentials
| Tethering Forces
| Tethering Forces
| Frozen Atoms
| Short Ranged (van der
| Short Ranged (van der
| Metal Potentials
| Three Body Potentials
| Four Body Potentials
| Direct Coulomb Sum
| Direct Coulomb Sum
| Truncated and Shifted Coulomb
| Truncated and Shifted Coulomb
| Coulomb Sum with Distance
| Coulomb Sum with Distance
| Ewald Sum
| Reaction Field
| Dynamical Shell Model
| Bond Constraints
| Bond Constraints
| Barostats
| Message 434: error -
| Message 434: error -
- sub-directory
- Macros
| cleanup
| cleanup
| copy
| select
| store
| store
| store
- bench
- The DL_POLY_2 Directory Structure
- build
- The DL_POLY_2 Directory Structure
- data
- The DL_POLY_2 Directory Structure
- execute
- The DL_POLY_2 Directory Structure
| Message 1: error -
- java
- The DL_POLY_2 Directory Structure
- public
- The DL_POLY_2 Directory Structure
- respa
- The DL_POLY_2 Directory Structure
- source
- The DL_POLY_2 Directory Structure
- utility
- The DL_POLY_2 Directory Structure
- thermostat
- Molecular Dynamics Algorithms
| External Fields
| Integration of the Rigid
| Integration of the Rigid
| The DL_POLY_2 Multiple Timestep
| The CONTROL File Directives
| Message 430: error -
| Message 464: error -
- Berendsen
- Berendsen Barostat
| Berendsen Barostat
| Integration of the Rigid
| Linked Rigid Bodies
| Test Case 4: Shell
| Test Case 5: Shell
- Nosé-Hoover
- The Hoover Barostat
| The Hoover Barostat
| Integration of the Rigid
| Linked Rigid Bodies
| Constructing Nonstandard Versions
| Test Case 3: An
- units
- DL_POLY
- Units
| Summary of Statistical Data
- energy
- .
- pressure
- Units
| Units
| The Hoover Barostat
| The CONTROL File Directives
| Pressure units:
| Summary of Statistical Data
- Verlet neighbour list
- Ewald Sum
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| The DL_POLY_2 Multiple Timestep
| Distributing the Nonbonded Terms
| Distributing the Nonbonded Terms
| Distributing the Nonbonded Terms
| Three Body Forces
| Metal Potentials
| Constructing Nonstandard Versions
| Constructing Nonstandard Versions
| Further Comments on the
| Further Comments on the
| Message 106: error -
| Message 107: error -
| Message 108: error -
| Message 109: error -
| Message 110: error -
- WWW
- The DL_POLY Package
| Obtaining the Source Code
| Obtaining the Source Code
- X-PLOR
- The DL_POLY Package
W Smith
2003-05-12
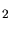 O
O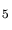 | Message 153: error -
| Message 385: error -
| Message 385: error -
| Message 386: error -
| Message 398: error -
| Message 422: error -
| Message 153: error -
| Message 385: error -
| Message 385: error -
| Message 386: error -
| Message 398: error -
| Message 422: error -
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
 T
T
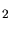 O
O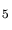 | Test Case 5: Shell
| Test Case 9: A
| Message 180: error -
| Message 185: error -
| Message 250: error -
| Message 330: error -
| Message 433: error -
| Test Case 5: Shell
| Test Case 9: A
| Message 180: error -
| Message 185: error -
| Message 250: error -
| Message 330: error -
| Message 433: error -
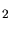 O
O | Test Case 3: An
| Test Case 9: A
| Message 383: error -
| Message 424: error -
| Message 446: error -
| Test Case 3: An
| Test Case 9: A
| Message 383: error -
| Message 424: error -
| Message 446: error -