



Next: Distributing the Intramolecular Bonded
Up: DL_POLY Parallelisation
Previous: DL_POLY Parallelisation
Contents
Index
The Replicated Data Strategy
The Replicated Data (RD) strategy [42] is one of several
ways to achieve parallelisation in MD.
Its name derives from the
replication of the configuration data on each node of a parallel
computer (i.e. the arrays defining the atomic coordinates
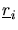 , velocities
, velocities
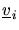 and forces
and forces
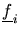 , for
all
, for
all 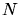 atoms
atoms
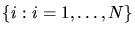 in the simulated system, are
reproduced on every processing node). In this strategy most of the
forces computation and integration of the equations of motion can be
shared easily and equally between nodes and to a large extent be
processed independently on each node. The method is relatively simple
to program and is reasonably efficient. Moreover, it can be
``collapsed'' to run on a single processor very easily. However the
strategy can be expensive in memory and have high communication
overheads, but overall it has proven to be successful over a wide
range of applications. These issues are explored in more detail in
[42,43].
in the simulated system, are
reproduced on every processing node). In this strategy most of the
forces computation and integration of the equations of motion can be
shared easily and equally between nodes and to a large extent be
processed independently on each node. The method is relatively simple
to program and is reasonably efficient. Moreover, it can be
``collapsed'' to run on a single processor very easily. However the
strategy can be expensive in memory and have high communication
overheads, but overall it has proven to be successful over a wide
range of applications. These issues are explored in more detail in
[42,43].
Systems containing complex molecules present several difficulties.
They often contain ionic species, which usually require Ewald summation methods [12,44], and intra-molecular
interactions in addition to inter-molecular forces. These are
handled easily in the RD strategy, though the SHAKE algorithm
[13] requires significant modification [30].
The RD strategy is applied to complex molecular systems as follows:
- Using the known atomic coordinates
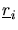 , each node
calculates
a subset of the forces acting between the atoms. These are usually
comprised of:
, each node
calculates
a subset of the forces acting between the atoms. These are usually
comprised of:
- atom-atom pair forces (e.g. Lennard Jones, Coulombic etc.);
- non-rigid atom-atom bonds;
- valence angle forces;
- dihedral angle forces;
- improper dihedral angle forces.
- The computed forces are accumulated in (incomplete) atomic
force arrays
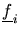 independently on each node;
independently on each node;
- The atomic force arrays are summed globally over all nodes;
- The complete force arrays are used to update the atomic
velocities and positions.
It is important to note that load balancing (i.e. equal and concurrent
use of all processors) is an essential requirement of the overall
algorithm. In DL_POLY_2 this is accomplished for the pair forces with an
adaptation of the Brode-Ahlrichs scheme [22].




Next: Distributing the Intramolecular Bonded
Up: DL_POLY Parallelisation
Previous: DL_POLY Parallelisation
Contents
Index
W Smith
2003-05-12