



Next: Distributing the Nonbonded Terms
Up: DL_POLY Parallelisation
Previous: The Replicated Data Strategy
Contents
Index
DL_POLY_2 handles the intramolecular
in which the atoms involved in any given bond term are explicitly
listed. Distribution of the forces calculations is accomplished by the
following scheme:
- Every atom in the simulated system is assigned a unique index
number from
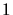 to
to 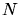 ;
;
- Every intramolecular bonded term
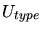 in the system has a
unique index number
in the system has a
unique index number 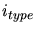 : from
: from 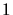 to
to 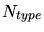 where
where 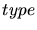 represents a bond, angle or dihedral.
represents a bond, angle or dihedral.
- A pointer array
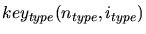 carries the
indices of the specific atoms involved in the potential term labelled
carries the
indices of the specific atoms involved in the potential term labelled
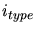 . The dimension
. The dimension 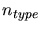 will be
will be 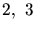 or
or 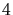 , if the
term represents a bond, angle or dihedral.
, if the
term represents a bond, angle or dihedral.
- The array
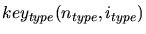 is used to identify
the atoms in a bonded term and the appropriate form of interaction and
thus to calculate the energy and forces. Each processor is assigned
the independent task of evaluating a block of
(
is used to identify
the atoms in a bonded term and the appropriate form of interaction and
thus to calculate the energy and forces. Each processor is assigned
the independent task of evaluating a block of
(
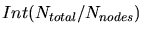 ) interactions.
) interactions.
The same scheme works for all types of bonded interactions. The
global summation of the force arrays does not occur until all the
force contributions, including nonbonded forces has been completed.




Next: Distributing the Nonbonded Terms
Up: DL_POLY Parallelisation
Previous: The Replicated Data Strategy
Contents
Index
W Smith
2003-05-12