![]()
In constructing a nonstandard DL_POLY_2 simulation program, the first requirement is for the user to write a program to function as the root segment. The source directory contains an example of such a root program: DLPOLY. This root program calls the major routines required to perform the simulation and also controls the normal ``molecular dynamics cycle'' which consists of forces calculation followed by integration of the equation of motion. DLPOLY also monitors the cpu usage and brings about a controlled termination of the program if the usage approaches the allotted job time within a pre-set closure time. Lastly, DLPOLY is the routine that first opens the OUTPUT file (section 4.2.2), which provides the summary of the job. Users are recommended to study the DLPOLY root as a model for other implementations of the package they may wish to construct. The dependencies and calling hierarchies of all the DL_POLY_2 subroutines can be found in the Appendix of the DL_POLY_2 Reference Manual.
If additional functionality is added to DL_POLY_2 by the user, the PARSET.F routine (and its support routines) will need modifying to allow specification of the dimensions of any new arrays. (A description of the existing arrays found in DL_POLY_2 is provided in the Appendix of the DL_POLY_2 Reference Manual.)
Any molecular dynamics simulation performs five different kinds of operation: initialisation; forces calculation; integration of the equations of motion; calculation of system properties; and job termination. It is worth considering these operations in turn and to indicate which DL_POLY_2 routines are available to perform them. We do not give a detailed description, but provide only a guide. Readers are recommended to examine the different routines described in chapter 8 of the DL_POLY_2 Reference Manual for further details (particularly regarding further dependencies i.e. additional routines that must be called.) The following outline assumes a system containing flexible molecules held together by rigid bonds, but without rigid bodies.
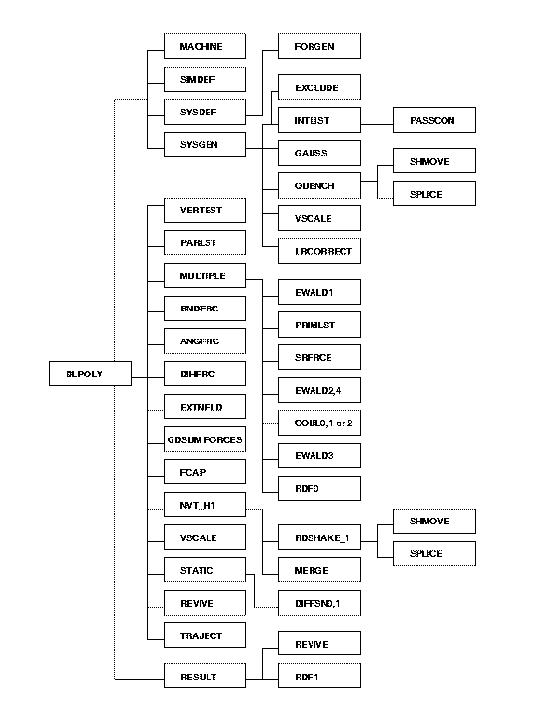
Initialisation requires firstly that the program determine what kind of parallel machine it is running on. The routine MACHINE determines how many processing nodes are being used and also returns the node identity to each process. Next the job control information is required; this is obtained by the routine SIMDEF, which reads the CONTROL file (section 4.1.1). The description of the system to be simulated: the types of atoms and molecules present and the intermolecular forces; are obtained by the SYSDEF routine, which reads the FIELD file (section 4.1.3). Lastly, the atomic positions and velocities must be provided. These are obtained by the SYSGEN routine, which reads the CONFIG file (section 4.1.2) and also generates the initial velocities if required to do so. If the system contains constraint bonds, the routine PASSCON is required to process molecular connectivity data and establish the communication procedure between nodes, and the QUENCH routine is required to set the starting velocities correctly. Also needed in the initialisation, is the routine FORGEN, which constructs the interpolation arrays for the short-range forces calculations, and the routine EXCLUDE which identifies atoms that are explictly chemically bonded through bonds, constraints or valence angles. The resulting list is known as the excluded atoms list.
The calculation of the pair forces represents the bulk of any simulation. A Verlet neighbour list is used by DL_POLY_2 in calculating the atomic forces. The routine that constructs this this is called PARLST. This routine builds the neighbour list taking into account the occurrence of atoms in the excluded atoms list. The routine SRFRCE calculates the short-range (van der Waals) forces, making use of the IMAGES routine to handle any periodic boundary conditions. Coulombic forces are handled by a varity of routines: COUL0, COUL1 and COUL2 handle Coulombic forces without periodic boundaries; EWALD1, EWALD2 and EWALD3 are used for systems with periodic boundaries (an additional routine: EWALD4 is necessary for the multiple timestep algorithm). Intramolecular forces require the routines ANGFRC, BNDFRC and DIHFRC. If the multiple timestep algorithm is required, the routine MULTIPLE must be used to call the various forces routines. It also calls the PRIMLST routine to split the interaction list into primary and secondary neighbours. The decision to update the neighbour list is handled by the routine VERTEST. The routine EXTNFLD is required if the simulated system has an external force field (e.g. electrostatic field) operating. To help with equilibration simulations, the routine FCAP is sometimes required to reduce the magnitude of badly equilibrated forces. Since DL_POLY_2 is based on the replicated data strategy, a global sum routine (GDSUM) is required to sum the atomic forces on all nodes.
Integration of the equations of motion is handled by one of the routines listed and described in section 2.5. For example routines NVE_0, NVT_E0, NVT_H0, NVT_B0 etc. are used if no constraint forces are present. These routines treat the NVE, Evans-NVT, Hoover-Nosť-NVT and NVT-Berendsen ensembles respectively. The corresponding versions of these routines which handle constraint forces are NVE_1, NVT_E1, NVT_H1 or NVT_B1. These versions call the routine RDSHAKE_1 to handle the constraints. RDSHAKE_1 itself calls a number of additional routines: MERGE, SHMOVE and SPLICE. For ad hoc temperature scaling, the routine VSCALEG is required.
As mentioned elsewhere, DL_POLY_2 does not contain many routines for computing system properties during a simulation. Radial distributions may be calculated however, using the routines RDF0 and RDF1. Similarly DIFFSN0 and DIFFSN1 calculate approximate mean square displacements. Ordinary thermodynamic quantities are calculated by the routine STATIC, which also writes the STATIS file (section 4.2.7). Routine TRAJECT writes the HISTORY (section 4.2.1) file for later analysis.
Job termination is handled by the routine RESULT which writes the final summaries in the OUTPUT file and dumps the restart files REVIVE and REVCON (sections 4.2.4 and 4.2.3 respectively).
An idea of the construction of a DL_POLY_2 program can be obtained from the following flowchart. The example represents a DL_POLY_2 program which uses the multiple timestep algorithm, with bond constraints and the Nosť-Hoover thermostat.
![]()